Metagenomics
There is growing interest in determining total species diversity in the environment, including in aquatic environments even in places as remote as the High Arctic. Only through a more complete profile of the living world can we truly understand how ecosystems function and adapt to changing conditions. Modern genomic sequencing has finally enabled us to generate these complete species profiles, from microorganisms all the way up to apex predators. This field is known as metagenomics.
We are now using metagenomic techniques to reach further into the dynamic and often murky worlds of marine and estuarine ecosystems. Our current focus is on the sub-tropical and temperate waters of the Indian River Lagoon on Florida's east coast, where changing environmental conditions have resulted in decreasing water quality and an increasing number of harmful algal blooms, or HABs, that threaten entire aquatic ecosystems and the health and livelihoods of a growing human population along its shores.
Our team is part of the Florida Center for Coastal and Human Health, a collaborative effort at HBOI that is conducting integrated research on the health of costal ecosystems and communities.

Krona representation of community structure
Currently, our collaborators and supporters include:


The low down on metagenomics
Metagenomic sequencing means that DNA from multiple species in an environment is collected and sequenced.
This can be accomplished by using all available DNA (whole genome shotgun), or by targeting a particular gene from all of the organisms present in the sample (16S, 18S).
This approach is used to determine the ‘community’ structure, which can capture additional species that may influence the survival of a particular species of interest.
Rapid detection of environmental DNA:
We have been developing the latest sequencing technology to collect information on the cohort of microorganisms involved during Florida’s period harmful algal blooms (HABs).
As part of the Florida Center for Coastal and Human Health, water samples from the Indian River Lagoon are used to obtain DNA for eukaryotic species.
What is Nanopore Sequencing?
Nanopore sequencing uses an electrical current to measure the passage of a DNA molecule through a small pore in a membrane. Each DNA base (G, A, T, C) makes a distinct change in the current as it passes through.
This technology is much more compact than other platforms, making it a popular choice for fieldwork.
Furthermore, the ability to produce data from a single molecule of DNA eliminates the the noise produced from methods relying on signal from a group of DNA molecules, allowing for much longer reads to be generated.


A common gene marker, 18S rRNA, is targeted and sequenced on the Oxford Nanopore MinION device.
This handheld sequencer is capable of producing real-time data with a minimal lab setup. The technology can be scaled to run samples from 12 sites at a time, or to run only a single site, for time-sensitive results that may address public health concerns related to HABs.
This small volume (11ul) contains enough DNA from ten different sites to identify hundreds of species using the MinION device.
During harmful algal blooms the presence of predator, prey, or competing species may greatly alter the course of a bloom –
thus we are not only interested in toxin-producing species, but also all of their neighbors.

The sequencing reaction is analyzed in real time, meaning that the raw data from each pore is being converted into sequence reads, quality filtered, and separated by barcode according to each site before the run is completed.


The DNA sequences are compared to a database of known microorganisms, and we can identify which species of phytoplankton are present. These data are compared across 20 sites over different times of the year.
All of the high quality DNA sequences from a single site are identified from a taxonomic database, allowing Krona (top) and Sankey (right) representations of their community structure.
Sankey representation of community structure
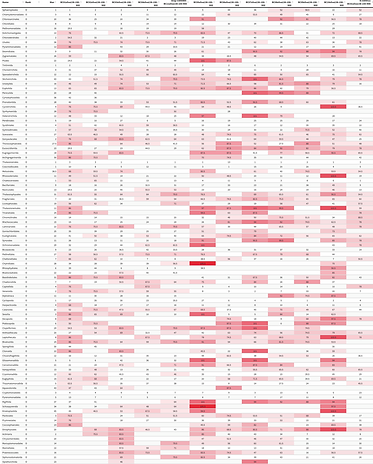
Heatmaps (left) are then generated to compare these communities across different sites and time periods.